Pulibications & Preprints
HIGHLIGHTS
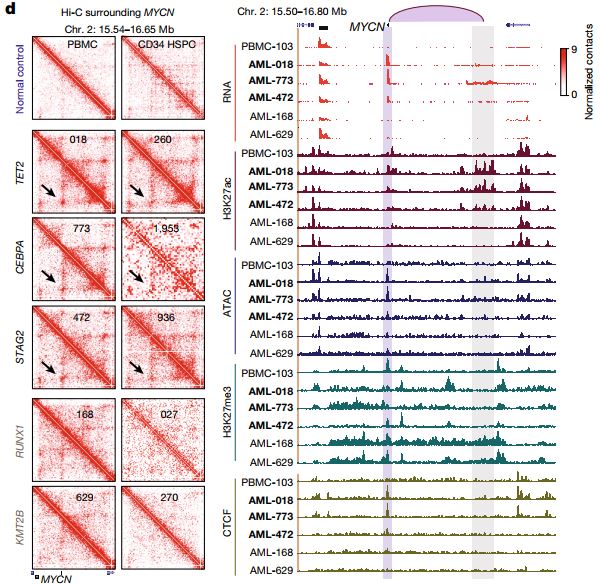
Subtype-specific 3D genome alteration in acute myeloid leukaemia
Nature
·
26 Oct 2022
·
doi:10.1038/s41586-022-05365-x
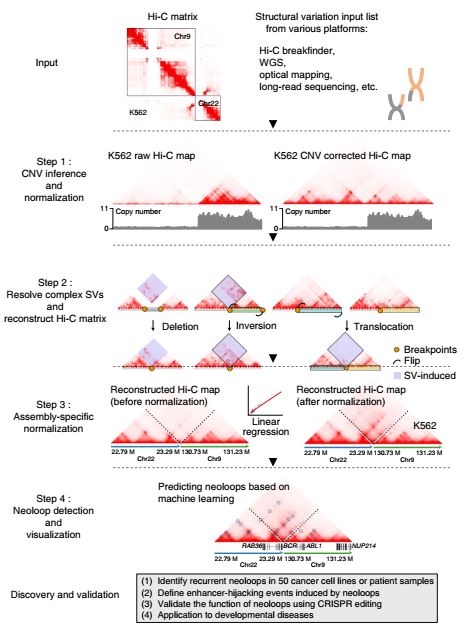
Genome-wide detection of enhancer-hijacking events from chromatin interaction data in rearranged genomes
Nature Methods
·
01 Jun 2021
·
doi:10.1038/s41592-021-01164-w
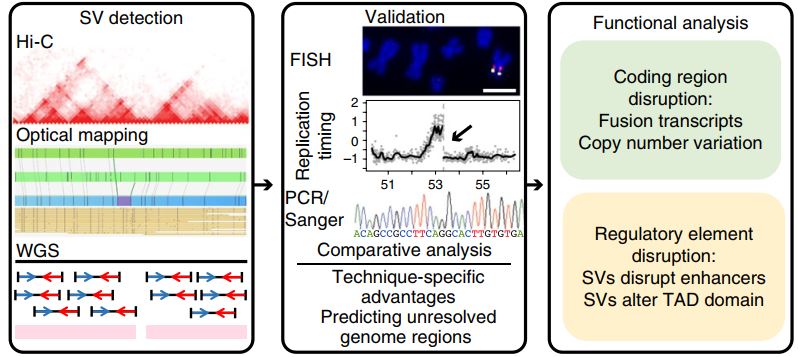
Integrative detection and analysis of structural variation in cancer genomes
Nature Genetics
·
10 Sep 2018
·
doi:10.1038/s41588-018-0195-8
LIST OF PUBLICATIONS
- J. Xu*, F. Song*, H. Lyu, M. Kobayashi, B. Zhang, Z. Zhao, Y. Hou, X. Wang, Y. Luan, B. Jia, L. Stasiak, J. H. Wong, Q. Wang, Qi Jin, Qiushi Jin, Y. Fu, H. Yang, R. C. Hardison, S. Dovat, L. C. Platanias, Y. Diao, Y. Yang, T. Yamada, A. Viny, R. L. Levine, D. Claxton, J. R. Broach, H. Zheng, F. Yue. Subtype-specific and reversible alteration of 3D genome structure in acute myeloid leukemia. Subtype-specific 3D genome alteration in acute myeloid leukaemia. Nature. 611, 387-398, doi:10.1038/s41586-022-05365-x (2022)
- J. Xu, B. Ren. Built to be imperfect. Nature Cell Biology 26 (8), 1229-1230 (2024).
- J. Dixon*, J. Xu*, V. Dileep*, Y. Zhan*, F. Song*, V. T. Le, G. G. Yardimci, A. Chakraborty, D. V. Bann, Y. Wang, R. Clark, L. Zhang, H. Yang, T. Liu, I. sriranga, L. An, C. Pool, T. Sasaki, J. C. R. Mulia, H. Ozadam, B. R. Lajoie, R. Kaul, M. Buckley, K. Lee, M. Diegel, D. Pezic, C. Ernst, S. Hadjur, D. T. Odom, J. A. Stamatoyannopoulos, J. R. Broach, R. Hardison, F. Ay, W. S. Noble, J. Dekker, D. M. Gilbert and F. Yue, An integrative detection and analysis of structural variation in cancer genomes, Nature Genetics. 50 (2018), no. 10, 1388-1398. (*co-first author)
- X. Wang, J. Xu, B. Zhang, Y. Hou, F. Song, H. Lyu, H. Yang, T. Liu, F. Yue. Genome-wide detection of enhancer-hijacking events from chromatin interaction data in rearranged genomes. Nature Methods. 18, 661-668, doi:10.1038/s41592-021-01164-w (2021).
- J. Zhang*, D. Lee*, V. Dhiman*, Peng Jiang*, J. Xu*, P. McGillivray*, H. Yang*, …, R. Klein, M. Snyder, D. M. Gilbert, K. Yin, C. Cheng, F. Yue, X. S. Liu, K. White and M. Gerstein. An integrative ENCODE resource for cancer genomics. Nature Communication. 11, 3696, (2020) (*co-first author)
- L. Li*, C. Hong*, J. Xu*, C. Y. Chung, A. K. Leung, D. A. Boncan, L. Cheng, K. Lo, P. Lai, J. Wong, J. Zhou, A. S. Cheng, T. Chan, F. Yue, K. Y. Yip. Accurate identification of structural variations from cancer samples. Briefings in Bioinformatics 25 (2023) (*co-first author)
- 4D Nucleome Consortium, J. Dekker, B. A. Oksuz, Y. Zhang, Y. Wang, M. K. Minsk, S. Kuang, L. Yang, J. H. Gibcus, N. Krietenstein, O. J. Rando, J. Xu … An integrated view of the structure and function of the human 4D nucleome. Nature (2025)
- L. Chang, Y. Xie, B. Taylor, Z. Wang, J. Sun, E. J Armand, S. Mishra, J. Xu, M. Tastemel, A. Lie, Z. A Gibbs, H. S Indralingam, T. M Tan, R. Bejar, C. C Chen, F. B Furnari, M. Hu, B. Ren. Droplet Hi-C enables scalable, single-cell profiling of chromatin architecture in heterogeneous tissues. Nature Biotechnology. 1-14 (Oct 2024).
- S. Zu, Y. E. Li, K. Wang, E. J. Armand, S. Mamde, M. L. Amaral, Y. Wang, A. Chu, Y. Xie, M. Miller, J. Xu, Z. Wang, K. Zhang, B. Jia, X. Hou, L. Lin, Q. Yang, S. Lee, B. Li, S. Kuan, H. Liu, J. Zhou, A. Pinto-Duarte, J. Lucero, J. Osteen, M. Nunn, K. A. Smith, B. Tasic, Z. Yao, H. Zeng, Z. Wang, J. Shang, M. M. Behrens, J. R. Ecker, A. Wang, S. Preissl, B. Ren. Single-cell analysis of chromatin accessibility in the adult mouse brain. Nature 624, no. 7991 (Dec 2023): 378-89.
- J. Xu, F. Song, E. Schleicher, C. Pool, D. Bann, M. Hennessy, K. Sheldon, E. Batchelder, C. Annageldiyev, A. Sharma, Y. Chang, A. Hastie, B. Miller, D. Claxton, G. Moldovan, F. Yue, J. R. Broach. An Integrated Framework for Genome Analysis Reveals Numerous Previously Unrecognizable Structural Variants in Leukemia Patients’ Samples. BioRXiv 563270. https://doi.org/10.1101/563270
- F. Song, J. Xu, J. Dixon. & Yue, F. Analysis of Hi-C Data for Discovery of Structural Variations in Cancer. Methods Mol Biol. 2301, 143-161, doi:10.1007/978-1-0716-1390-0_7 (2022).
- H. Yang, H. Zhang, Y. Luan, T. Liu, W. Yang, K. G. Roberts, M. Qian, B. Zhang, W. Yang, V. Perez-Andreu, J. Xu, S. Iyyanki, D. Kuang, L. Stasiak, S. C. Reshmi, J. Gastier-Foster, C. Smith, C. Pui, W. E. Evans, S. P. Hunger, L. C. Platanias, M. V Relling, C. G. Mullighan, M. L Loh, F. Yue, J. Yang. Noncoding genetic variation in GATA3 increases acute lymphoblastic leukemia risk through local and global changes in chromatin conformation. Nature Genetics. 54, 170-179 (2022)
- T. Iyyanki, B. Zhang, Q. Wang, Y. Hou, Q. Jin, J. Xu, H. Yang, T. Liu, X. Wang, F. Song, Y. Luan, H. Yamashita, R. Chien, H. Lvu, L. Wang, J. Warrick, J. Raman, J. Meeks, D. DeGraff, F. Yue. Subtype-associated epigenomic landscape and 3D genome structure in bladder cancer. Genome Biology 22:105, (2021).
- H. Yang, Y. Luan, T. Liu, H. Lee, L. Fang, Y. Wang, X. Wang, B. Zhang, Q Jin, K. Ang, X. Xing, J. Wang, J. Xu, F. Song, I. Sriranga, C. Khunsriraksakul, T. Salameh, D. Li, M. Choudhary, J. Topczewski, K. Wang, G. Gerhard, R. Hardison, T. Wang, K. Cheng, F. Yue. A map of cis-regulatory elements and 3D genome structures in zebrafish. Nature. 588, 337-343 (2020)
- The ENCODE Project Consortium. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature. 583, 699–710 (2020)
- Z. Yan, N. Huang, W. Wu, W. Chen, Y. Jiang, J. Chen, X. Huang, X. Wen, J. Xu, Q. Jin, K. Zhang, Z. Chen, S. Chien, S. Zhong. Genome-wide colocalization of RNA–DNA interactions and fusion RNA pairs. Proceedings of the National Academy of Sciences. 2019 116 (8) 3328-3337.
- Y. Wang, F. Song, B. Zhang, L. Zhang, L. An, J. Xu, D. Li, M. N. Choudhary, Y. Li, M. Hu, R. Hardison, T. Wang and F. Yue, The 3D Genome Browser: A web-based browser for visualizing 3D genome organization and long-range chromatin interactions, Genome Biology. 19 (2018), no. 1, 151.
- F. Xian, J. Xu, L. Nakhleh. Detecting large indels using optical map data. RECOMB International conference on Comparative Genomics. 2018. vol 11183.
- L. Jiang, M. Yin, J. Xu, M. Jia, S. Sun, X. Wang, J. Zhang, D. Meng. The Transcription Factor Bach1 Suppresses the Developmental Angiogenesis of Zebrafish. Oxid Med Cell Longev, 2017.
- R. Fu, R. J. Gill, E. Y. Kim, N. E. Briley, E. R. Tyndall, J. Xu, C. Li, K. S. Ramamurthi, J. M. Flanagan, F. Tian, Spherical nanoparticle supported lipid bilayers for the structural study of membrane geometry-sensitive molecules, Journal of the American Chemical Society. 137 (2015), no. 44, 14031-14034.
- L. Jiang, M. Yin, X. Wei, J. Liu, X. Wang, C. Niu, X. Kang, J. Xu, Z. Zhou, S. Sun, X. Wang, X. Zheng, S. Duan, K. Yao, R. Qian, N. Sun, A. Chen, R. Wang, J. Zhang, S. Chen, D. Meng. Bach1 represses Wnt/β-catenin signaling and angiogenesis. Circulation Research. 2015;117:364–375
- J. Xu, J. Liu, L. Jiang, D. Meng. Roles of reactive oxygen species in pluripotent stem cell functions. Chinese Journal of Pathophysiology, 2013.